In the same section
- Polytech
- 3BIO
- BioInfo
- Thesis and Intership subjects
-
Share this page
Master Thesis subjects proposed by 3BIO-BioInfo Computational Biology and Bioinformatics
Our lab
Computational biology and bioinformatics is a field that permeates through all levels of science, from fundamental research to technology development and application. Our lab focuses on the development of bioinformatic tools and computational approaches that can be used both for investigating fundamental questions in biology and designing practical solutions particularly in biotechnology and medicine.
The methods used to build these tools are varied but mainly revolve around artificial intelligence, machine learning, data analysis, statistical physics, stochastic modelling and quantum chemistry. Experimental data is of paramount importance for most of these methods and we have therefore established strong connections with experimental labs from both academia and the industry. Moreover, our lab is part of a network of collaborators within the ULB-VUB (IB)2 Interuniversity Institute of Bioinformatics in Brussels.
The bulk of our work consists in developing a suite of tools that enables the prediction of the changes caused by mutations on the biophysics of proteins (stability, solubility, binding affinity, …) and DNA (ionisation potential). These tools can be applied to study fundamental questions such as the identification and modelling of the mechanisms of natural selection, as well as bringing solutions to the design and optimisation of enzymes used in biotechnological processes or the identification of deleterious variants in diseases like cancer and autism.
Furthermore, we are developing tools for the prediction of epitopes in antigens, which can be applied to vaccine and antibody design, and the understanding of the mechanisms behind certain cancers, auto-immune diseases and allergies.
Finally, we are also working on the mathematical modelling of complex biological systems in order to gain insight, for example, on the role of noisy interactions in systems like genetic circuits or chemical reaction networks or to study how known physiological pathways are perturbed in infection and diseases.
.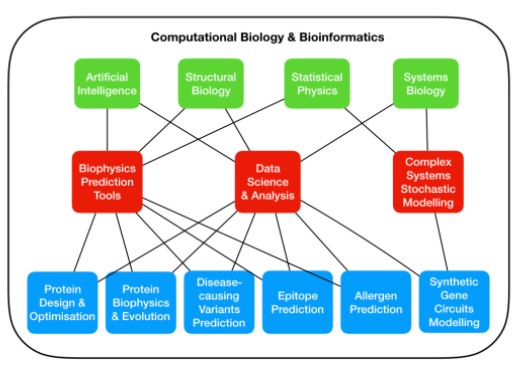
Schematic representation of our lab’s topics of research (blue) according to the methods used (red) and their relevant fields of study (green).
Topics
1. Rational design of modified proteins [ Fabrizio Pucci & Marianne Rooman ]
Proteins carry out a large variety of biological functions in living organisms. Some proteins, for example, act as particularly specific and efficient catalysts. The possibility to exploit the functional properties of proteins in industrial applications (food industry, chemical processes, pharmaceutical developments, etc.) is extremely interesting; however, one of its major limitations is that proteins generally lose their stability and activity under non-physiological conditions. The ability to design modified proteins that remain structured, soluble and active at higher (or lower) temperatures is therefore an important research objective. Moreover, the understanding of how changes in the coding sequence of proteins affect their biophysical properties is essential in fundamental research such as molecular evolution and phylogeny. This project includes the development of efficient and fast computational tools to predict changes in stability, solubility or interactions of proteins upon mutations, which are applicable on a genome-wide scale. Examples of algorithms already developed for protein design can be found at dezyme.com. The new software constructed in the framework of the master thesis will be applicable to the rational modification of proteins of industrial interest and to the increase of their efficiency in a given process, or to study fundamental questions of interest in biophysics and molecular evolution. This project will be done in our group or in collaboration with experimental laboratories in academia or industry where the predictions will be experimentally tested.
2. Prediction of disease-causing variants in the human genome [ Fabrizio Pucci & Marianne Rooman ]
Next Generation Sequencing technologies produce massive amounts of genome data that are revolutionizing biological and medical research, and paves the way towards personalized medicine. Among the exome variants that lead to amino acid mutations, most are neutral in the sense that they only modify the individual’s phenotype, but some are the cause of diseases. The identification of deleterious mutations and their characterization are of prime importance for setting up personalized therapies. This project consists of developing and applying bioinformatics tools to predict disease-causing protein variants and trying to understand why they are so, in terms of protein characteristics such as stability, solubility, flexibility and function, or in terms of DNA characteristics such as its ionisation potential. We recently developed FiTMuSiC, a tool combining evolutionary and structural information to predict variant fitness, which was one of the top performance methods in the Critical Assessment of Genome Interpretation competition. We are currently applying it to characterize, at the molecular level, mutations involved in rare Mendelian diseases, skin cancer and neurodevelopmental disorders such as autism spectrum disorder and Alzheimer. The project is in collaboration with geneticists and clinicians (e.g., Hôpital Erasme, BE, Dana Farber Cancer Institute, USA, University of California San Diego School of Medicine, USA).
3. Monoclonal Antibody design [Fabrizio Pucci & Marianne Rooman]
Maintaining global health requires the development of generic and versatile technologies that allow fast and effective responses to the large variety of disorders, in particular cancer and emerging infectious diseases.
Among these, monoclonal antibodies (mAbs) play an important role. Indeed, antibodies can bind antigens, such as bacterial or viral proteins or proteins expressed in cancer cells, and trigger the human immune response. In this project, we will build a bioinformatics pipeline for the design of mAbs that can bind with high affinity a specific target antigen. For that purpose, we will rely on experimentally characterized antibody-antigen complexes and their binding affinity values, detect informative sequence- and structure-based features, and combine them into a predictor using artificial intelligence techniques. The mAbs that we will design will be tested in vitro by experimental collaborators. While the project is focused on the design of a generic pipeline, it can be applied to specific case studies such as chronic lymphocytic leukemia, on which we are currently collaborating with cancer immunologists (Institute Bordet, BE), or COVID in collaboration with the CER research center.
4. Mathematical modeling of the Renin-Angiotensin System [Fabrizio Pucci & Marianne Rooman]
The renin-angiotensin system (RAS) is an important pathway that plays a key role in many physiological functions among which inflammation and blood pressure regulation. Its dysregulation is related to multiple pathological conditions such as hypertension and acute respiratory distress syndrome, just to mention some of them. In this project, we will use the ordinary differential equation formalism to study and predict the evolution of RAS under different perturbations. We previously analyzed the effect of SARS-CoV-2 on RAS and how the administration of an endogenous peptide could rescue the severity of the disease. Now we are extending the RAS model to other diseases such as sepsis, septic shock and cardiogenic shock, as clinical data are available. This project is in collaboration with the Intensive Care Unit of the Hôpital Erasme.
5. Artificial intelligence methods to design ligands for olfactory receptors [Dimitri Gilis]
The olfactory system relies on protein receptors expressed by olfactory neurons. These olfactory receptors belong to the family of G protein-coupled membrane receptors (GPCR). The relationships between odorant molecules, targeted olfactory receptors and odour perception are complex and not yet well understood. In addition, it has been shown that some olfactory receptors are expressed in tissues other than the olfactory epithelium and may have a physiological or potentially therapeutic role.
This project consists in developing artificial intelligence approaches, allowing (1) to predict the olfactory receptor(s) targeted by an odorant molecule, and (2) to design de novo a molecule able to activate a given olfactory receptor. It is carried out in collaboration with the group of Prof. I. Langer (Faculty of Medicine), which experimentally characterises these systems.
The master thesis topics related to this project can be entirely bioinformatics or include an experimental part.
6. Food and house dust mite allergens [Dimitri Gilis]
Allergy represents an important public health problem. On the one hand, we are developing bioinformatics tools to predict whether a protein corresponds to a food allergen. Such tools are very important for the development of new food products. On the other hand, we are studying certain structural and dynamic properties of house dust mite allergens.
7. Leveraging deep learning model for biomolecule [Fabrizio Pucci & Marianne Rooman & Dimitri Gilis]
Deep Learning algorithms are revolutionizing biomolecular science. For example, the AlphaFold2 and RosettaTR algorithms recently succeeded in pushing the accuracy of 3D protein structure prediction close to that of experimental methods. In this line of research, we will explore complex deep learning architectures to predict targeted biophysical characteristics of proteins. We propose two subtopics. In the first, pretrained attention-based language models (LM), such as ProteinBert, will be used and fine-tuned on downstream tasks such as protein localization or melting temperature predictions. Depending on the availability of computational resources, we will also consider direct training of BERT-like models which use masked LM approaches. The second subtopic is focused on the exploration of generative models for protein design.
.PDF File
Contacts
Pr. Dimitri Gilis: dgilis@ulb.ac.bePr. Fabrizio Pucci: fapucci@ulb.ac.be
Pr. Marianne Rooman : mrooman@ulb.ac.be
